Adjust Clustering Parameters
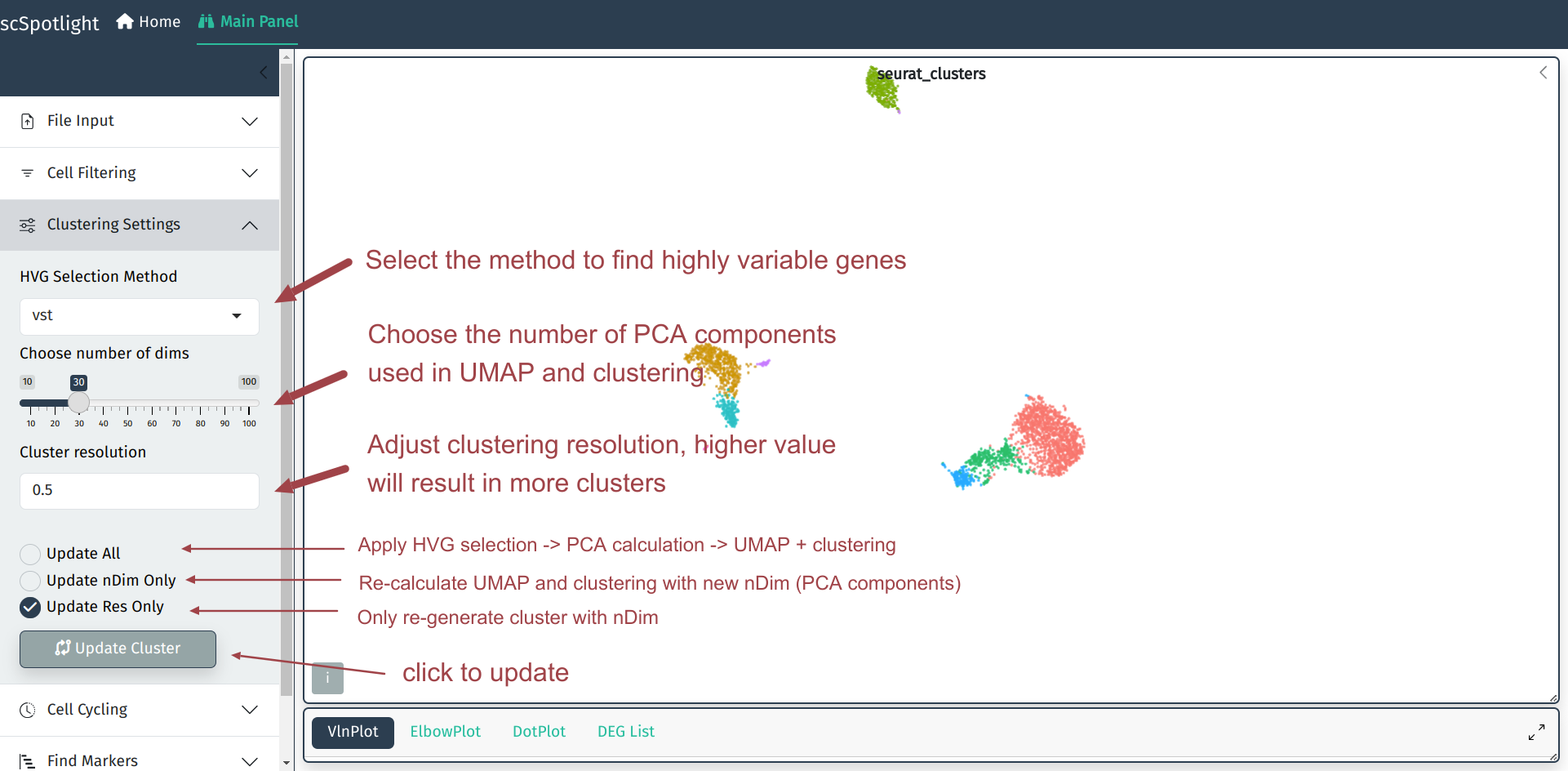
In most cases, the default parameters for clustering will not always satisfy user’s requirements. scSpotlight allows users to easily choose the method of highly variable genes selection, tune dimension reduction and clustering arguments on the left side bar, thus better identify the rare cell types.
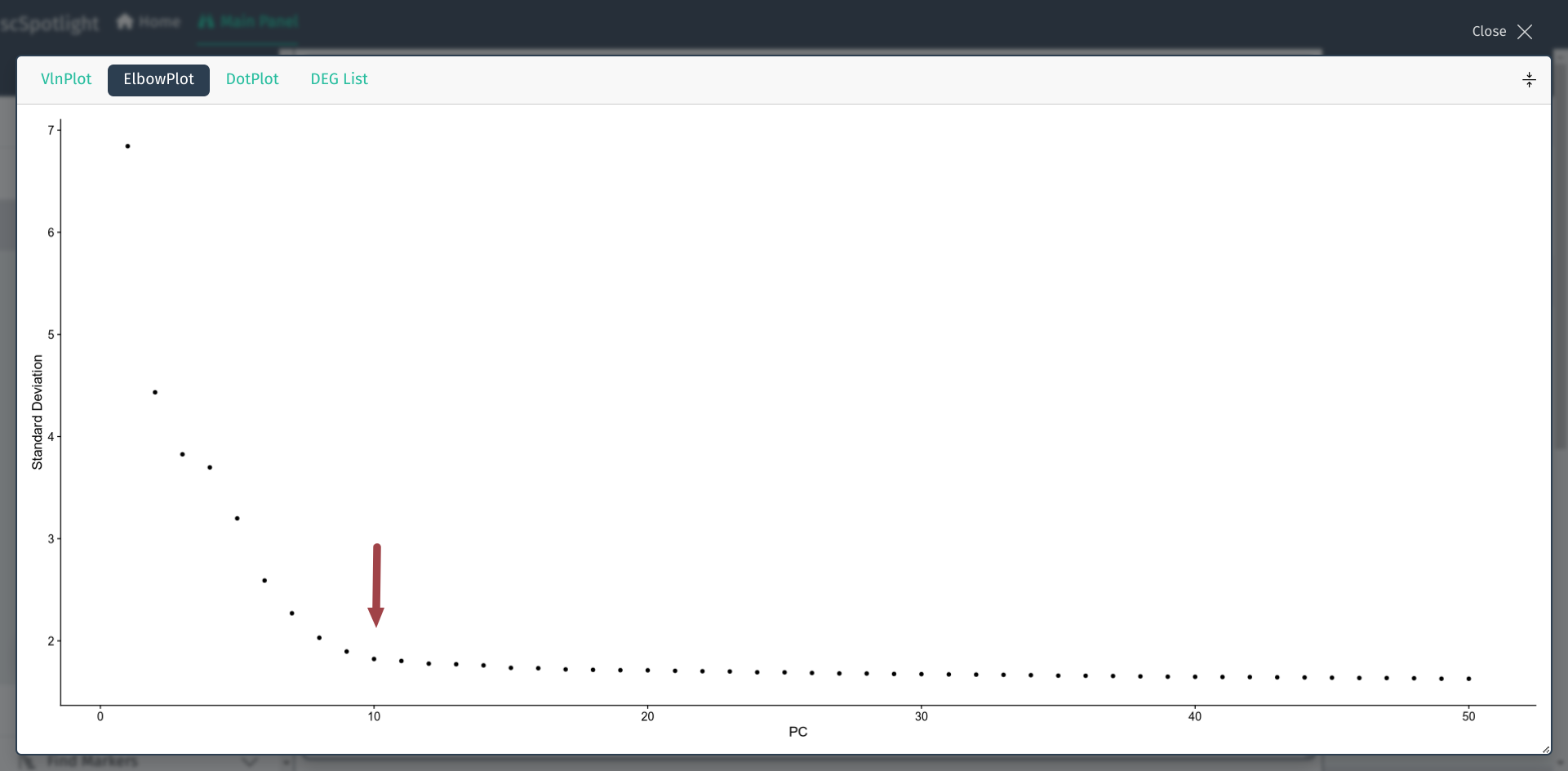
In dimension reduction step, user will have to choose an optimized number of PCA components as the input of non-linear dimension reduction (UMAP/TSNE). This value could be selected according to elbow plot in Seurat analysis workflow. User could expand the information panel to view elbow plot, Y axis is the percentage of the variance that each component could explain, X axis is the ranked components. Typically, user could select the “elbow” point. For instance, in Seurat pbmc3k tutorial, they used nDim=10.
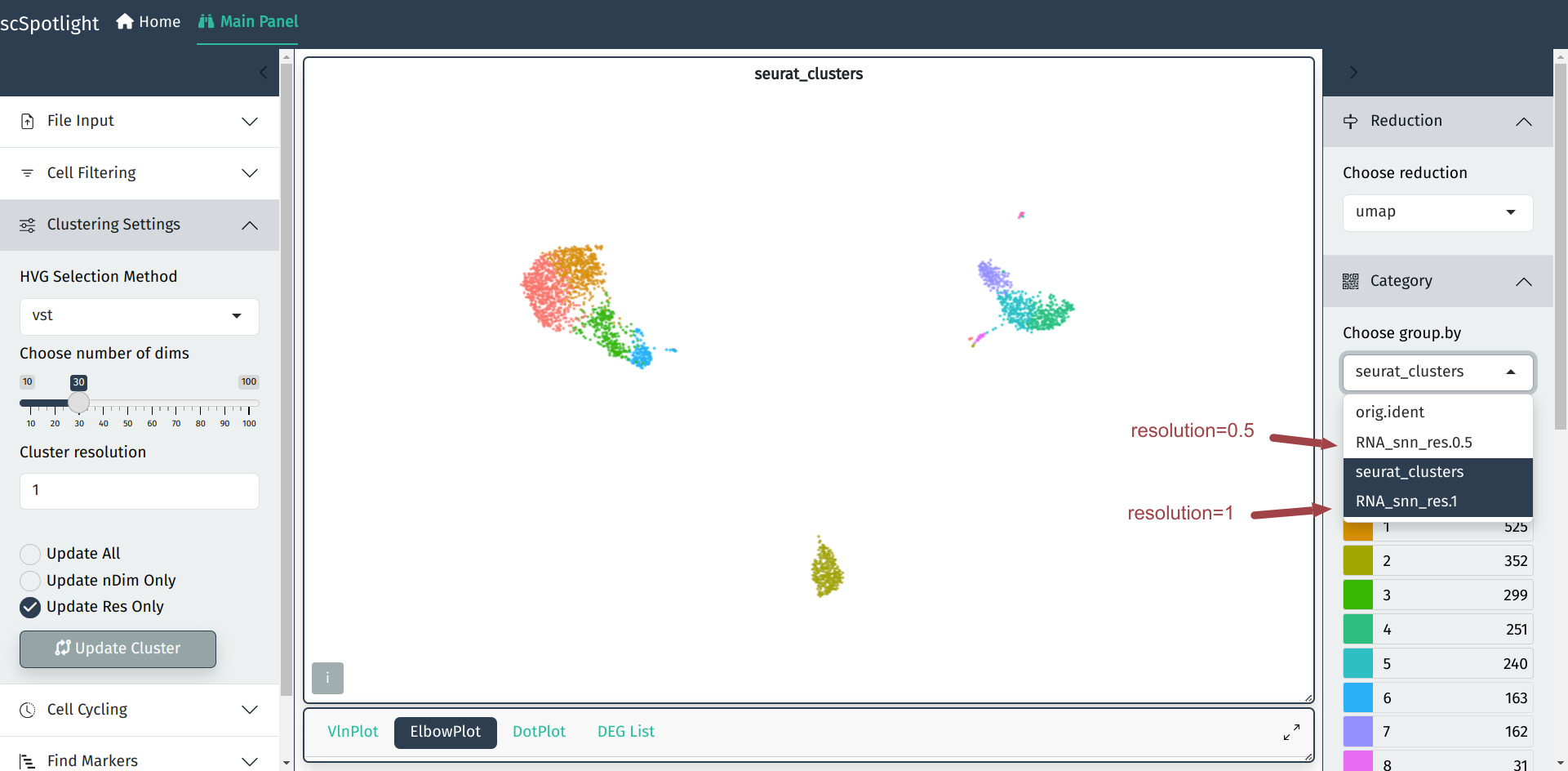
After finding cell clusters with different resolution, the updated grouping information could be chosen from the right-side panel.